
Gene/protein expression and regulation
Developmental and tissue specific transcriptome, post-transcriptional regulation and proteome data provide essential information to informatively translate a pathogen’s genome. Synchronized life cycle stages and dissected individual tissues are used to successfully compartmentalize the functions and define phylogenetic conservation and diversification among species.

Evolution of genomes, genes and proteins
Using comparative analysis and evolutionary principles we have discovered key insight into the unexpectedly rapid pace of new protein evolution within a metazoan and particularly within the Nematoda and Platyhelminthes Phyla. We furthermore focus on protein family birth and death, and protein expansion and reductions over evolutionary time.

Vaccine targets
We use a reverse vaccinology approach (based on protein conservation, gene expression profiles over the life cycles, MS proteomic detection, host antibody production to nematode targets) to identified nematode-originated molecules that activate innate and acquired immunity that we are exploring as promising targets for broad spectrum anthelmintic vaccine targets.
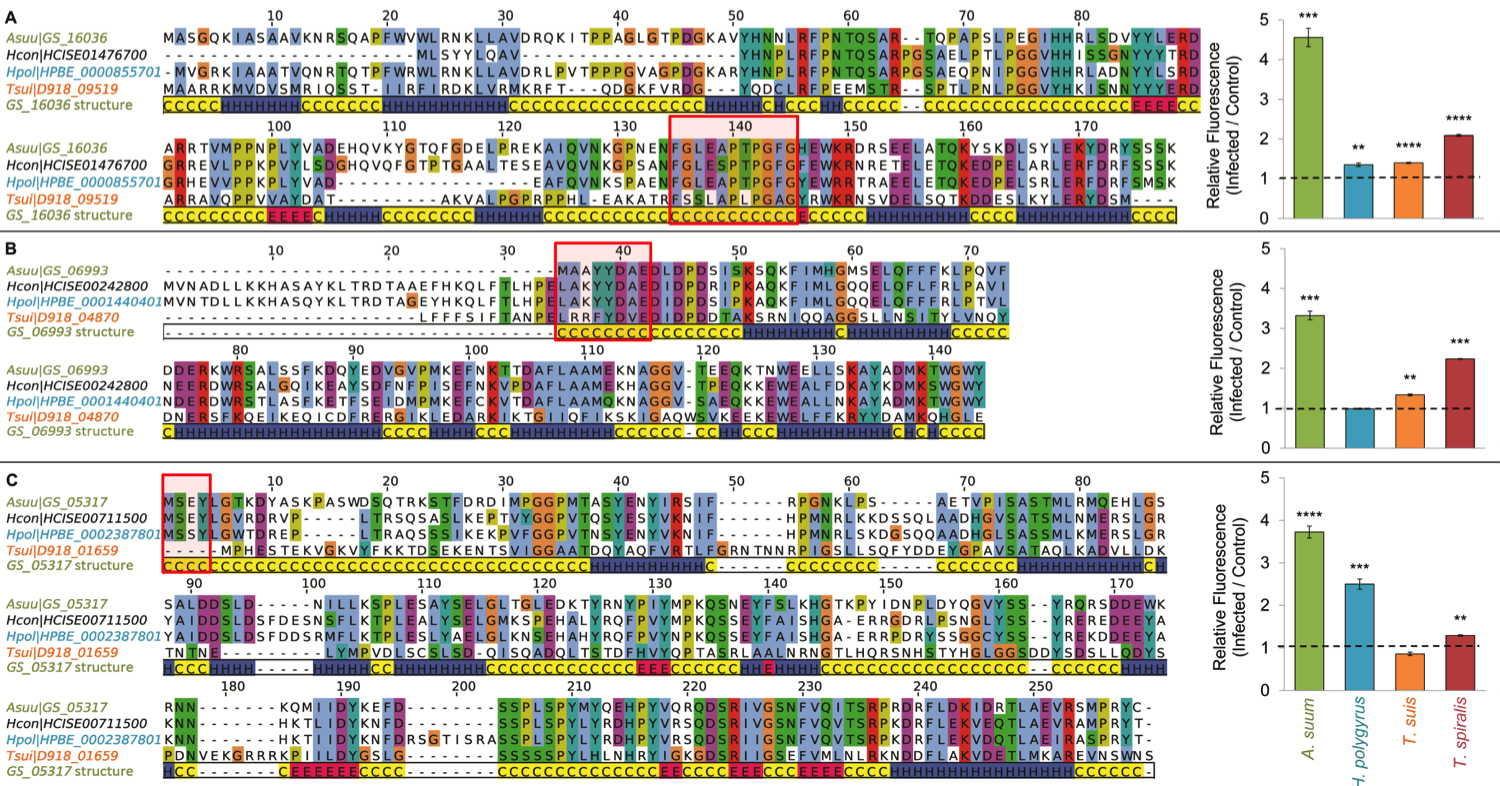
Diagnostics
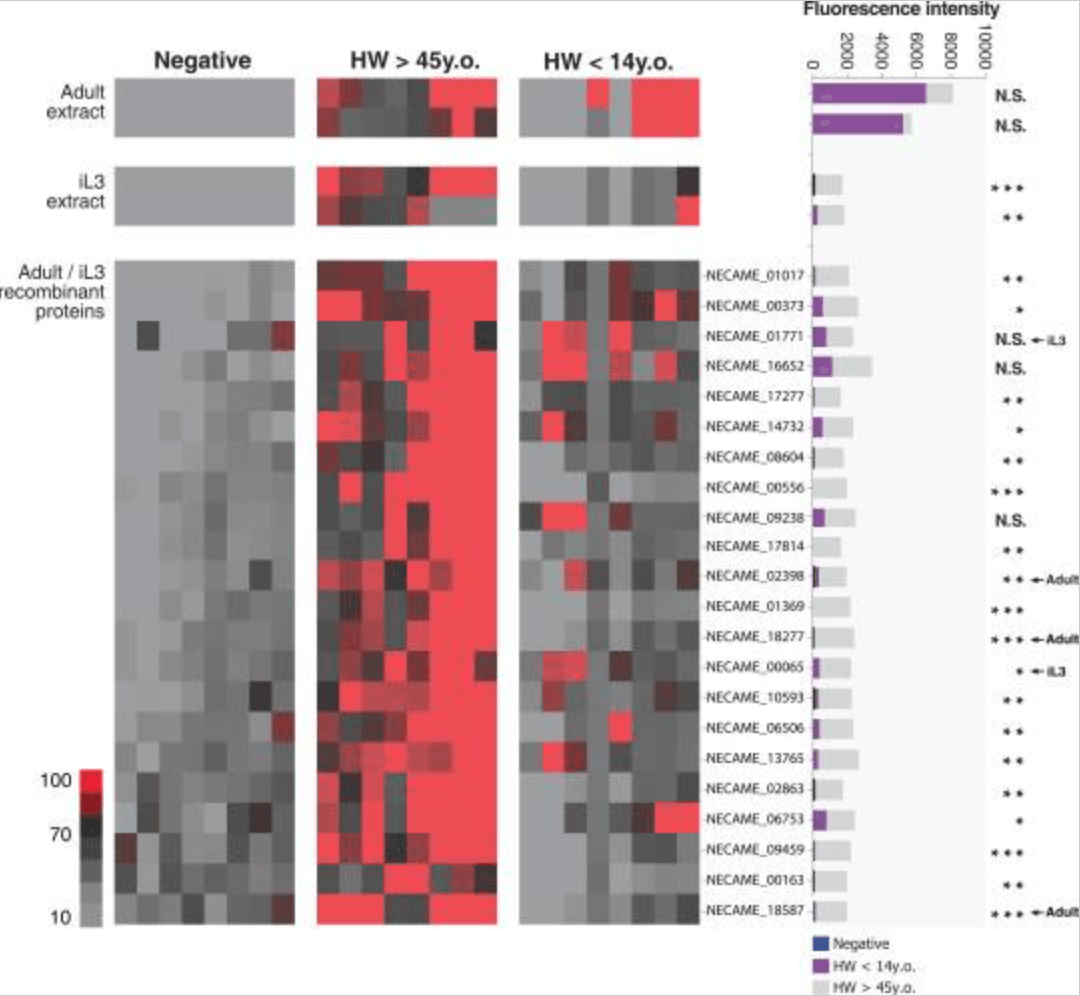
Host response to infection
We analyze high-throughput host transcriptomics (RNA-Seq) and proteomics (protein microarray) to identify genes and functional pathways altered in response to infections. Analyzing host response (human and mice) using lung tissues, PBMCs, T cells, B cells to a variety of infections (helminths, tuberculosis, malaria), provides better understanding of the infection process and the immune response.

Discovery of drug targets and drugs

Population genomics and drug resistance
We study genome variations within and between populations to gain insights into geographical differentiation and gene flow, transmission patterns and evolution of parasites. We use GWAS and forward genetic screens that can greatly facilitate identification of genetic variants correlated with phenotypes of biomedical interests (e.g., infection behavior, drug resistance, etc.), central to understanding population biology and molecular epidemiology of helminth parasites.

Microbiome in infectious diseases
The human intestine and its microbiota is the most common infection site for soil-transmitted helminths, therefore we study the complex cross-kingdom interactions between worm, host immune system and the micro biome. We use targeted and shotgun metagenomics to determine changes in micro biome assemblages during and after clearance of infection.
Databases, bioinformatic approaches & tools
We have a sustained commitment to go beyond the generation of sequence data. Through the development of public facing databases we organize our data in a manner that can be easily used by the scientific community in general. We also generate tools and approaches for the analysis of high-throughput parasite omics data.