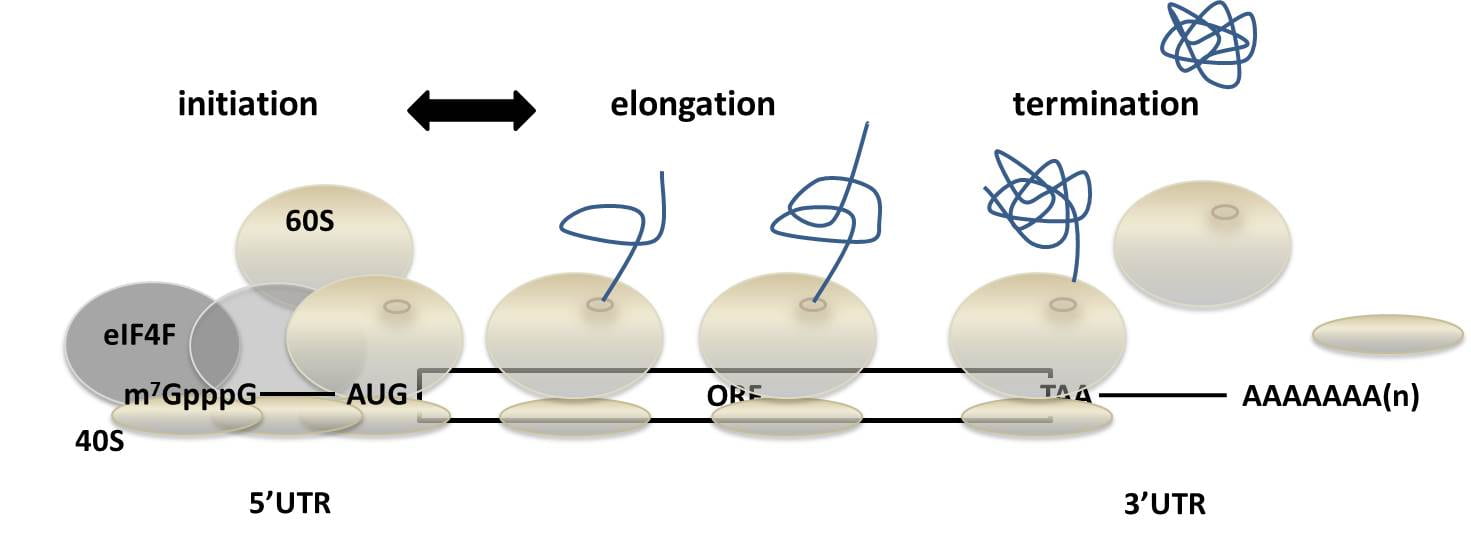
Our interest in sequence determinants of efficient protein synthesis has been focused on the transition from translation initiation to translation elongation and the so-called translational ramp (Verma et al., 2019). Djuranovic lab has shown that translational efficiency is regulated by amino-acid sequence composition as well as local mRNA sequence at the very N-terminus or the beginning of the coding sequence. Following translation kinetics, we indicated substantial ribosomal pausing and abortion of protein synthesis (peptidyl drop-off) on the 4th or 5th codon for distinct amino acid or nucleotide compositions. Our first findings in E. coli cells and in in vitro translation systems are now extended to eukaryotic systems (mainly, yeast and human). We were able to determine parts of the ribosome sequence and structure that are responsible for this selection on the nascent polypeptide synthesis. Our future directions on this research topic will focus on evolutionary and disease-related aspects of our findings. In this respect, evolutionarily, we are interested in organism-specific ribosome selection on the first five amino acids in endogenous gene sequences and for disease models we are following a set of disease-related genes with enriched mutations in the first five codons of the coding sequence as well as changes in ribosomal sequences/modifications in the peptide exit channel. We are also interested in the developing biotechnological tools that increase production of recombinant proteins.
