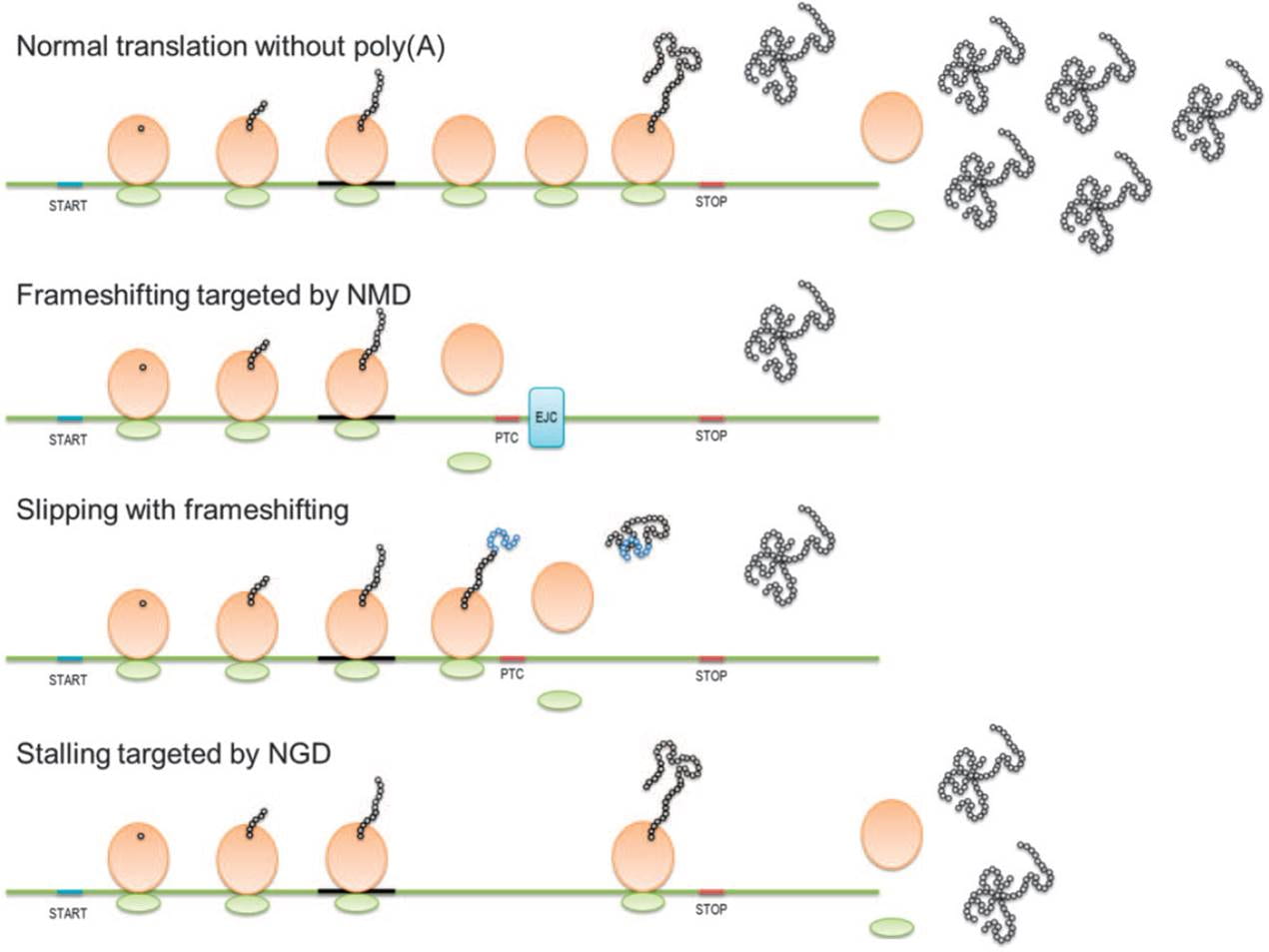
We are also interested in mRNA sequence determinants of translation efficiency that govern differences in both translation initiation and elongation (Arthur et al., 2015, 2017, 2018, Koutmou et al., 2015, Habich et al, 2016, Pavolvic Djuranovic et al, 2019, Verma et al., 2019). We established the polyA track motif, defined as an mRNA motif with 12 consecutive adenosine nucleotides in the coding sequence of mRNA, as a translational attenuator in both prokaryotic and eukaryotic organisms (Arthur et al., 2015 and Koutmou et al. 2015, reviewed in Arthur et al., 2018). We discovered that the presence of polyA track motifs in an mRNA sequence induces ribosomes to stall and misread the encoded information (Arthur et al., 2015, 2017, 2018, Koutmou et al., 2015, Habich et al., 2016). We indicated that these sequence motifs are present in in 2% of genes in most eukaryotic genomes (Habich et al., 2016).
A large number of genes with endogenous coding polyA motifs are conserved in major biological processes such as rRNA and mRNA biogenesis, general control of transcription, and the cell cycle. Having abnormal levels of a particular protein or any non-natural, frameshifted protein may negatively impact the health of the cell and could ultimately lead to disease. As such, we are focusing on silent mutations within polyA track motifs that were long considered to be innocuous but may disrupt the intricate balance that governs gene product levels. Our goal is to dissect the biology of polyA track motifs to broaden our understanding of gene regulatory pathways and cell homeostasis, as well as the impact of silent mutations in these homo-polymeric coding regions. Since polyA tracks are able to regulate gene expression by functioning as a “gene expression restraint” (Arthur et al., 2015), we used polyA tracks to develop a tool to create hypomorphic alleles of reporter and endogenous genes in multiple species (Arthur et al., 2017). This biotechnological tool is now patented (PCT/US2017/041766) and licensed to Canopy Biosciences as TUNR Flexible Gene Editing system.
