Dissect the molecular mechanisms of selective neuronal and regional vulnerability in Lewy body diseases (LBDs) using single-cell multi-omics
A fundamental characteristic of all neurodegenerative diseases is the pathology associated with the disease only affects particular neurons (‘selective neuronal vulnerability’) in particular brain regions (‘selective regional vulnerability’). The discovery of key pathways that regulate this differential susceptibility of neurons to degeneration holds great potential for the discovery of novel drug targets and the development of promising neuroprotective treatment strategies. However, the mechanisms underlying selective neuronal vulnerability have been difficult to dissect because of our limited ability to distinguish different neuronal subpopulations. Single cell/nucleus technology is an unbiased approach well suited for identifying and characterizing distinct cell populations in the brains.
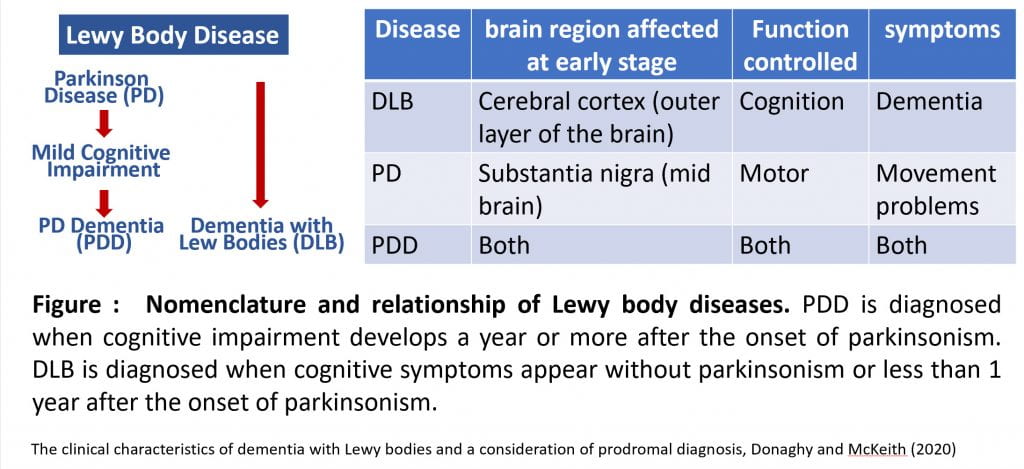
Lewy body diseases (LBDs) include Parkinson disease (PD), Parkinson disease dementia (PDD) and dementia with Lewy bodies (DLB). The three disorders can be viewed as existing on a spectrum of Lewy body disease because they share clinical, pathological, genetic and biochemical signatures. However, they have distinctive Lewy body deposition patterns that correlate with neuronal losses and clinical manifestations unique to each disease. We will generate a single-cell multi-omic atlas of different human brain regions from patients with Lewy body disease or cognitively normal controls. We will determine cell-type-specific transcriptomic and epigenomic changes for each cell type in the brain in each disease condition. By integrating multi-omic data, genomics, epigenomics and machine learning and analyzing disease-perturbed gene networks we will identify biomarkers to distinguish these closely related diseases and candidate targets for therapeutic treatment development.