Based on the order, directionality, and number of elongation cycles involved in peptide maturation, nonribosomal peptide (NRP) biosynthesis can be classified as linear, iterative, or nonlinear. In linear nonribosomal peptide synthetase (NRPS) assembly, the number and sequence of incorporated substrates in the final product are corresponding to the number and order of modules in the NRPS. The straightforward sequence of reactions results in continuous elongation of the peptide chain until a termination event where the product is released from the NRPS assembly line. This results in a linear relationship between NRPS domain counts and the molecular weight of the synthesized nonribosomal peptide. However, there are instances where this relationship deviates from the expected trend. Such deviations can be attributed to different factors. Above the linear trend line, points may represent NRPSs that employ redundant modules. Points below the linear trend line indicate 1) NRPSs utilize iterative or nonlinear assembly logic, 2) the gene cluster is not complete, or 3) additional enzyme machinery is employed for substrate incorporation. By comparing the NRPS domain counts and the molecular weight of synthesized NRP, we can give a rough prediction for the type of NRPSs (linear VS nonlinear).
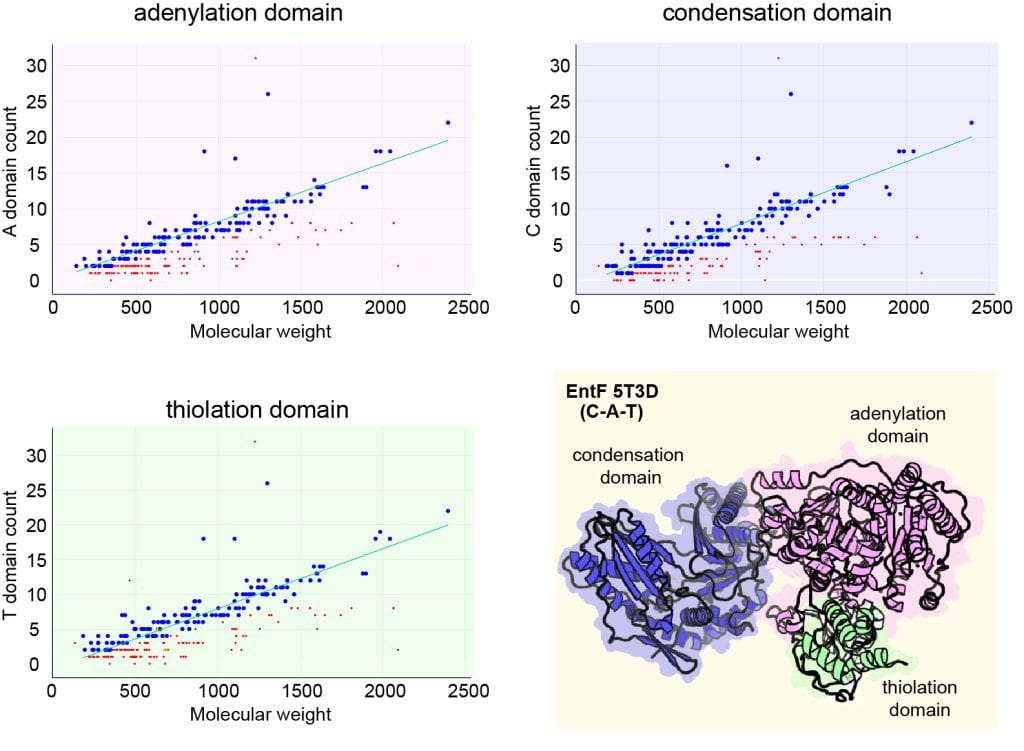
- NRPS prediction based on adenylation domain count
- NRPS prediction based on condensation domain count
- NRPS prediction based on thiolation domain count
Database source: MIBiG database
Coding package for NRPS-Typer: download here