42. Kim E., Dai B., Qiao J.B., Li W., Fortner J.D., Zhang F., Microbially Synthesized Repeats of Mussel Foot Protein Display Enhanced Underwater Adhesion. ACS Appl. Mater. Interfaces, 10 (49), 43003–43012 (2018) DOI: 10.1021/acsami.8b14890.
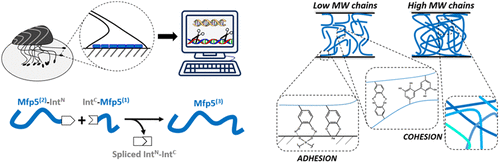
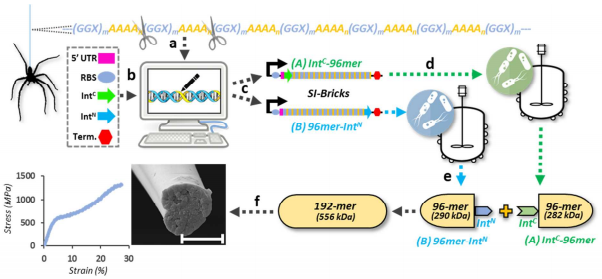
41. Bowen C.H.*, Dai B.*, Sargent C.J., Bai W, Ladiwala P, Feng H., Huang W, Kaplan D.L., Galazka J.M., Zhang F., Recombinant Spidroins Replicate Mechanical Properties of Natural Spider Silk. Biomacromolecules, 19 (9), 3853–3860 (2018) DOI: 10.1021/acs.biomac.8b00980. *These authors contributed equally.
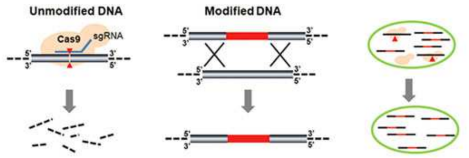
40. Xiao Y, Wang S, Rommelfanger S, Balassy A, Barba-Ostria C, Gu P, Galazka JM, Zhang F. Developing a Cas9-based tool to engineer native plasmids in Synechocystis sp. PCC 6803. Biotechnol Bioeng. Jun 13. (2018) doi: 10.1002/bit.26747.
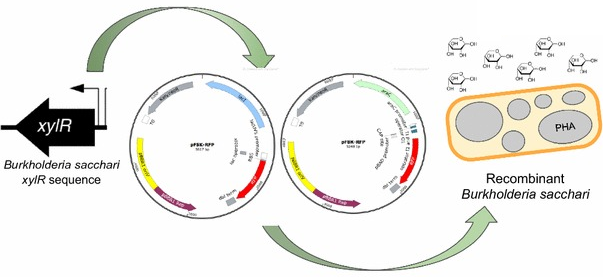
39. Guamán LP*, Barba-Ostria C*, Zhang F, Oliveira-Filho ER, Gomez JGC, Silva LF. Engineering xylose metabolism for production of polyhydroxybutyrate in the non-model bacterium Burkholderia sacchari. Microb Cell Fact. 17(1):74. (2018) doi: 10.1186/s12934-018-0924-9.*These authors contributed equally.
38. Liu D, Mannan AA, Han Y, Oyarzún DA, Zhang F, Dynamic metabolic control: towards precision engineering of metabolism. J Ind Microbiol Biotechnol. (2018). doi: 10.1007/s10295-018-2013-9
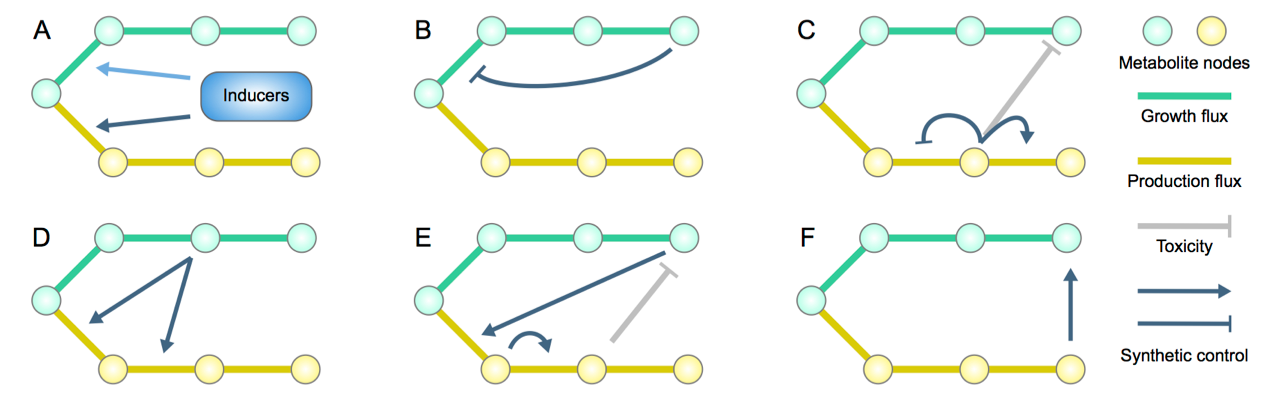
37. Liu D, Zhang F., Metabolic Feedback Circuits Provide Rapid Control of Metabolite Dynamics. ACS Syn Biol. 7 (2), 347–356, doi: 10.1021/acssynbio.7b00342 (2018).
36. Jiang W, Gu P, Zhang F., Steps towards ‘drop-in’ biofuels: focusing on metabolic pathways. Current Opinion in Biotechnology. 53, 26-32 (2018) doi.org/10.1016/j.copbio.2017.10.010.
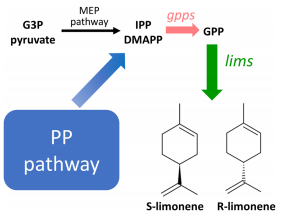
35. Lin PC, Saha R, Zhang F, Pakrasi HB., Metabolic engineering of the pentose phosphate pathway for enhanced limonene production in the cyanobacterium Synechocystis sp. PCC 6803. Scientific Reports. 7(1), 17503. doi: 10.1038/s41598-017-17831-y. (2017).
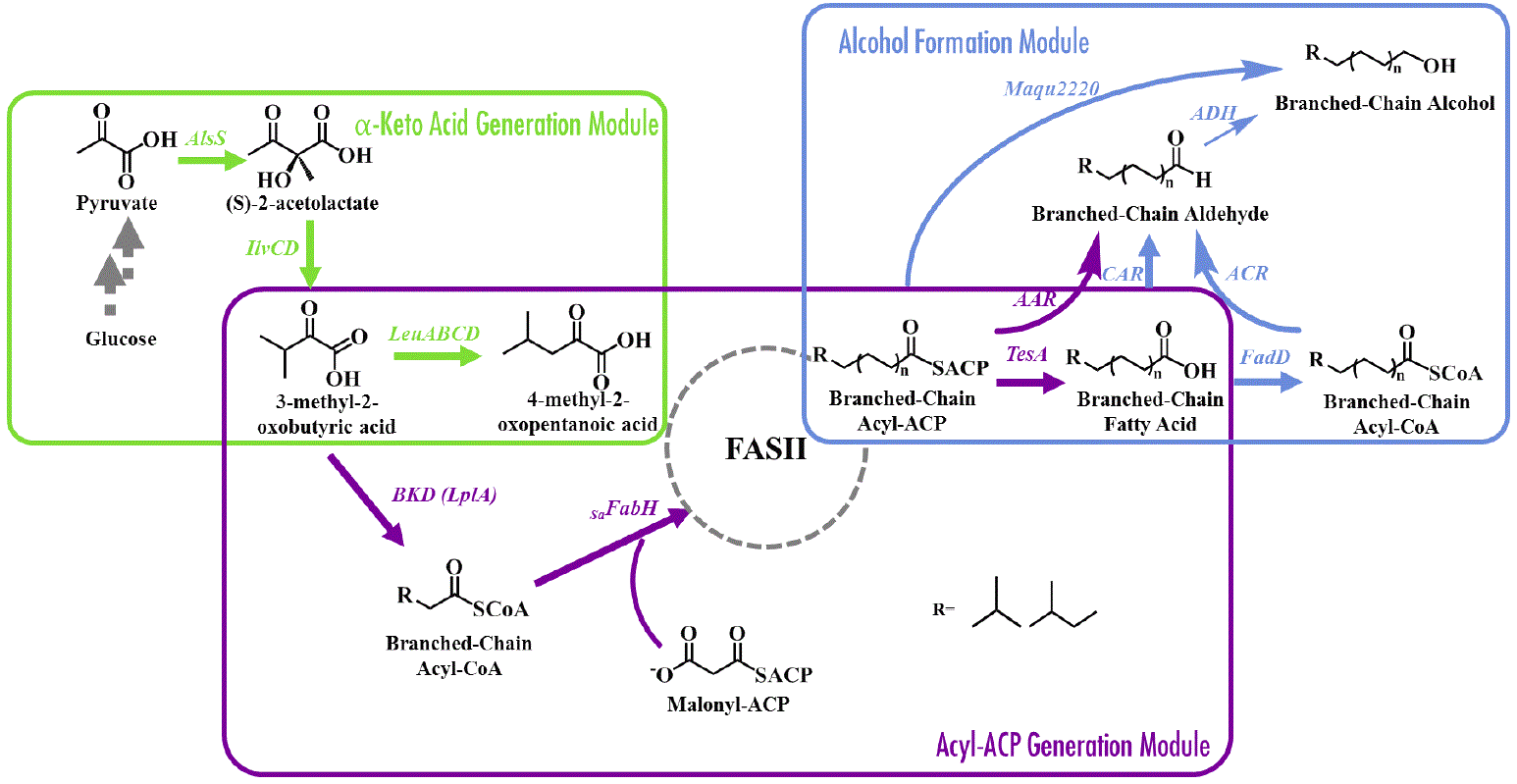
34. Jiang W., Qiao J.B., Bentley G.J., Liu D., Zhang F., Modular pathway engineering for the Production of Branched-Chain Fatty Alcohols. Biotechnol Biofuels. 10:244. doi: 10.1186/s13068-017-0936-4. eCollection (2017).
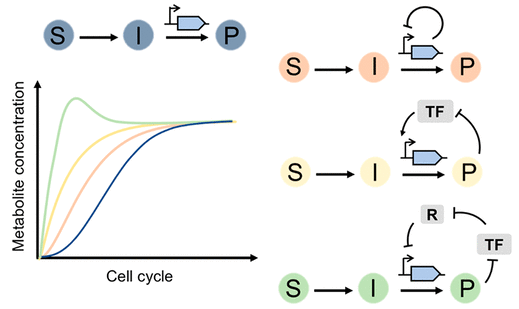
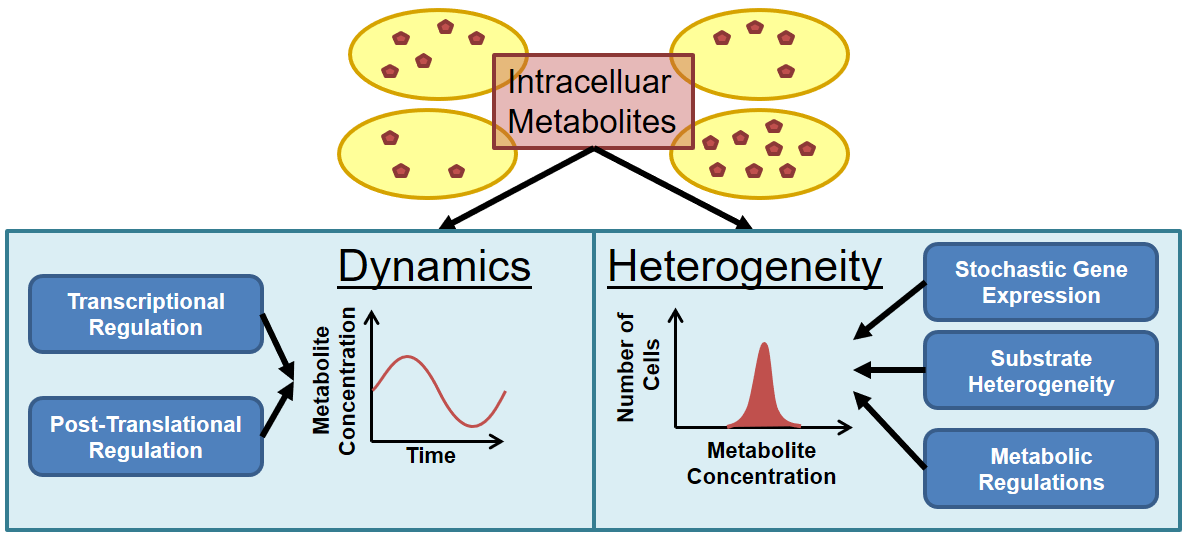
33. Schmitz AC, Hartline CJ, Zhang F, Engineering Microbial Metabolite Dynamics and Hetergeneity. Biotech J . ASAP, (2017) Sep 13. doi: 10.1002/biot.201700422.
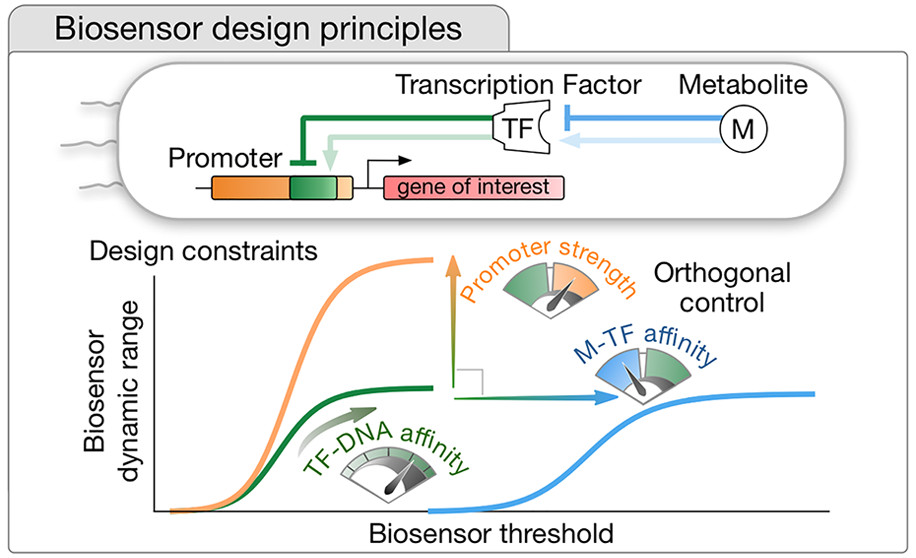
32. Mannan AA, Liu D, Zhang F, Oyarzún DA., Fundamental Design Principles for Transcription-Factor-Based Metabolite Biosensors.ACS Syn Biol. 6(10):1851-1859. doi: 10.1021/acssynbio.7b00172, (2017).
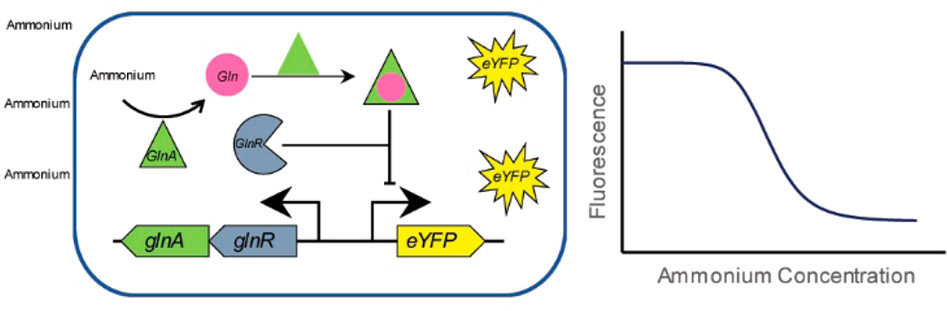
31. Xiao Y, Jiang W, Zhang F., Developing a Genetically Encoded, Cross-Species Biosensor for Detecting Ammonium and Regulating Biosynthesis of Cyanophycin. ACS Syn Biol. 6(10):1807-1815. doi: 10.1021/acssynbio.7b00069. (2017).
30. Liu D., Wan N., Zhang F., Tang Y.J., Wu S.G., Enhancing fatty acid production in Escherichia coli by Vitreoscilla hemoglobin overexpression. Biotechnol Bioeng. 114, 463, (2017).