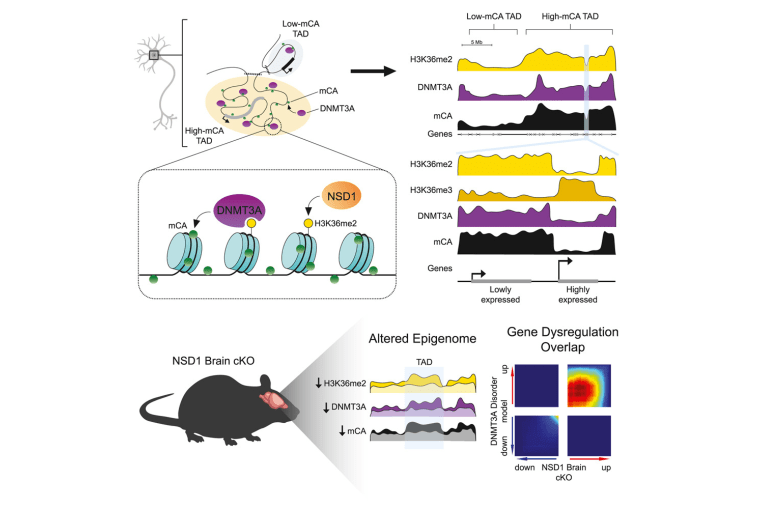
NSD1 deposits histone H3 lysine 36 dimethylation to pattern non-CG DNA methylation in neurons
Hamagami N, Wu DY, Clemens AW, Nettles SA, Li A, Gabel HW. NSD1 deposits histone H3 lysine 36 dimethylation to pattern non-CG DNA methylation in neurons. Molecular Cell. 2023; 83(9):1412-1428.e7. https://doi.org/10.1016/j.molcel.2023.04.001
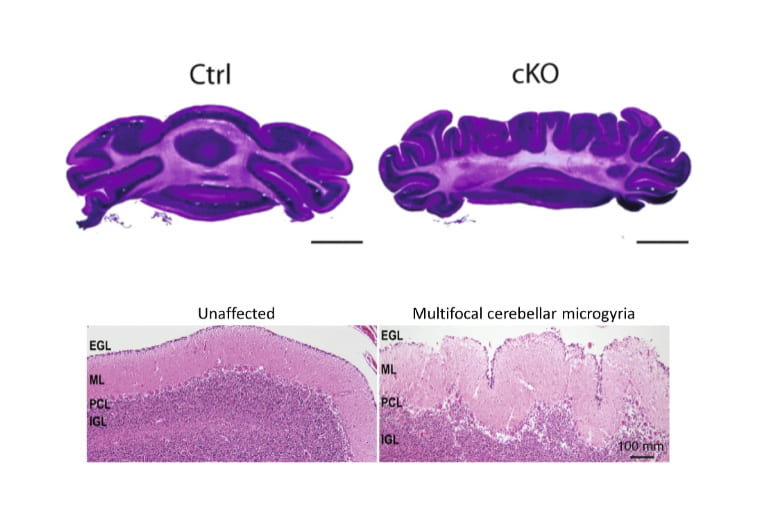
CHARGE syndrome protein CHD7 regulates epigenomic activation of enhancers in granule cell precursors and gyrification of the cerebellum
Reddy NC, Majidi SP, Kong L, Nemera M, Ferguson CJ, Moore, M, Gocalves TM, Liu H-K Fitzpatrick JAJ, Zhao G, Yamada T, Bonni A, Gabel HW. Nature Communications. 2023; 12:5702.
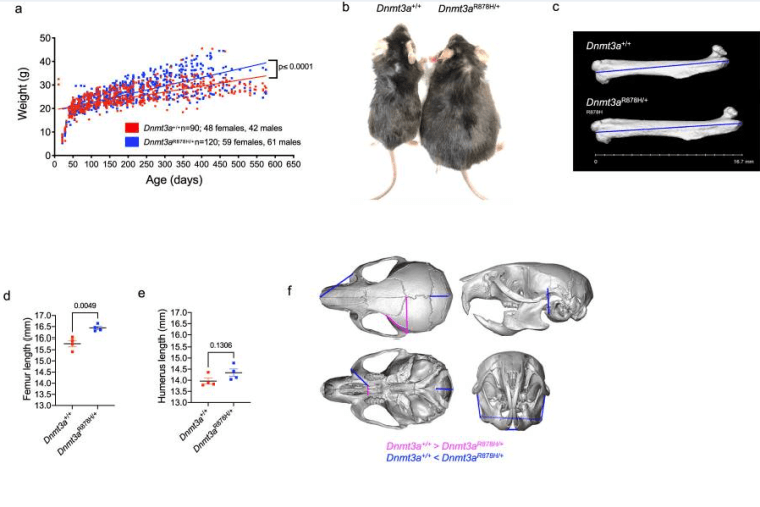
Functional and epigenetic phenotypes of humans and mice with DNMT3A Overgrowth Syndrome
Smith AM, LaValle TA, Shinawi M, Ramakrishnan SM, Abel HJ, Hill CA, Kirkland NM, Rettig MP, Helton NM, Heath SE, Ferraro F, Chen DY, Adak S, Semenkovich CF, Christian DL, Martin JR, Gabel HW, Miller CA, Ley TJ. Nat Commun. 2021 Jul 27;12(1):4549. doi: 10.1038/s41467-021-24800-7.
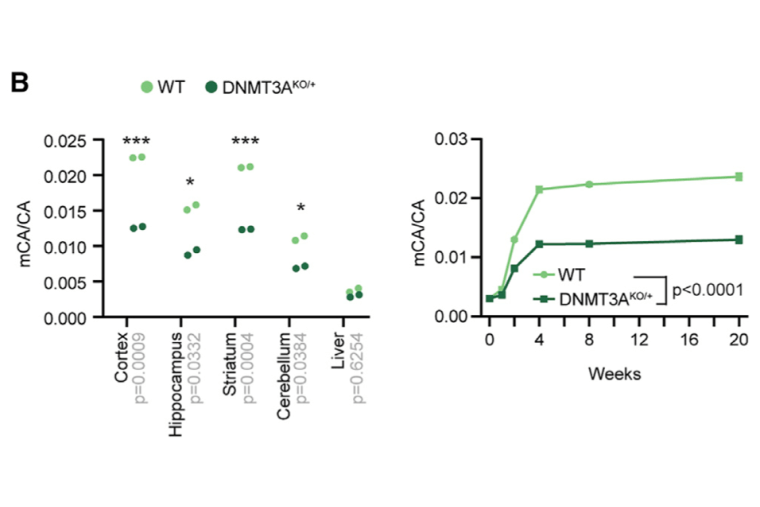
DNMT3A haploinsufficiency results in behavioral deficits and global epigenomic dysregulation shared across neurodevelopmental disorders
Christian DL*, Wu DY*, Martin JR, Moore JR, Liu YR, Clemens AW, Nettles SA, Kirkland NM, Hill CA, Wozniak DF, Dougherty JD, Gabel HW. Cell Reports. 2020 Nov 24;33(8):108416. doi: 10.1016/j.celrep.2020.108416.
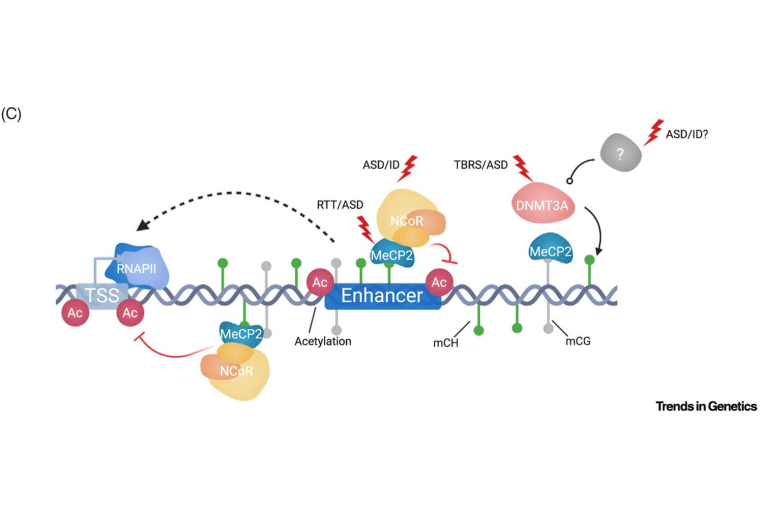
Emerging insights into the distinctive neuronal methylome
Clemens AW, Gabel HW. Trends in Genetics. 21 August 2020. doi: https://doi.org/10.1016/j.tig.2020.07.009
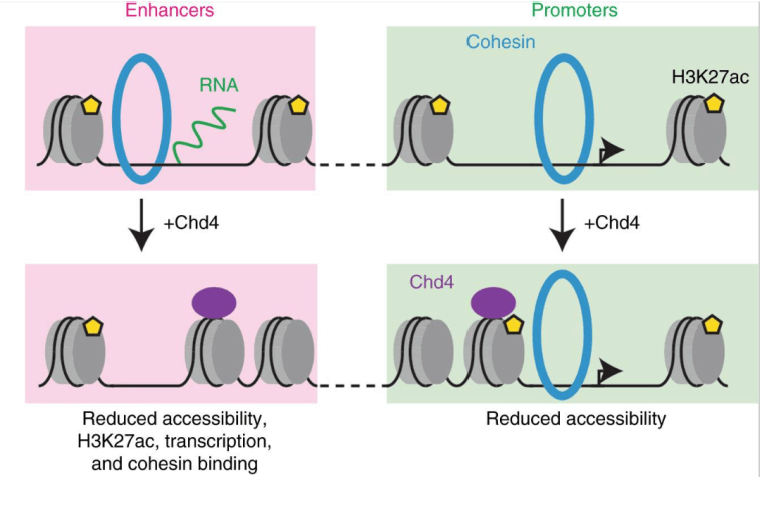
The chromatin remodeling enzyme Chd4 regulates genome architecture in the mouse brain
Goodman JV, Yamada T, Yang Y, Kong L, Wu DY, Zhao G, Gabel HW, Bonni A. Nature Communications. 9 July 2020. https://doi.org/10.1038/s41467-020-17065-z
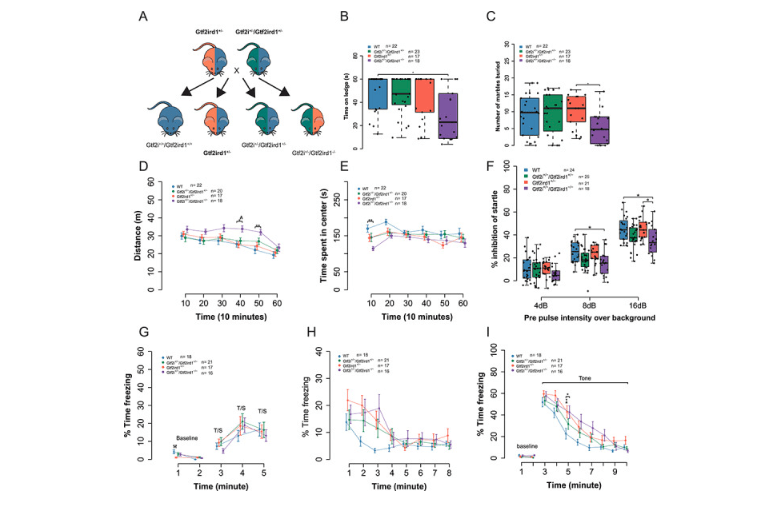
Functions of Gtf2i and Gtf2ird1 in the developing brain: transcription, DNA binding and long-term behavioral consequences
Kopp ND, Nygaard KR, Liu Y, McCullough KB, Maloney SE, Gabel HW, Dougherty JD. Human Molecular Genetics. 1 May 2020. https://doi.org/10.1093/hmg/ddaa070
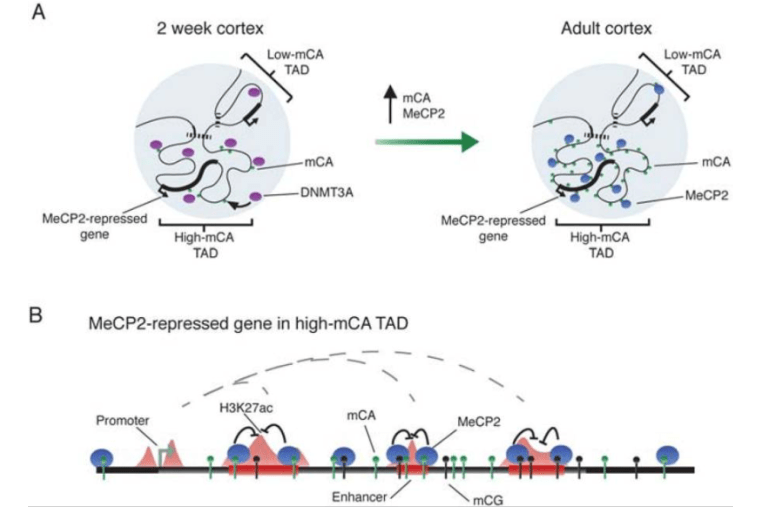
MeCP2 represses enhancers through chromosome topology-associated DNA methylation
Clemens AW*, Wu DY*, Moore JR, Christian DL, Zhao G, Gabel HW, MeCP2. Mol Cell. 2019 Nov 26. doi: 10.1016/j.molcel.2019.10.033
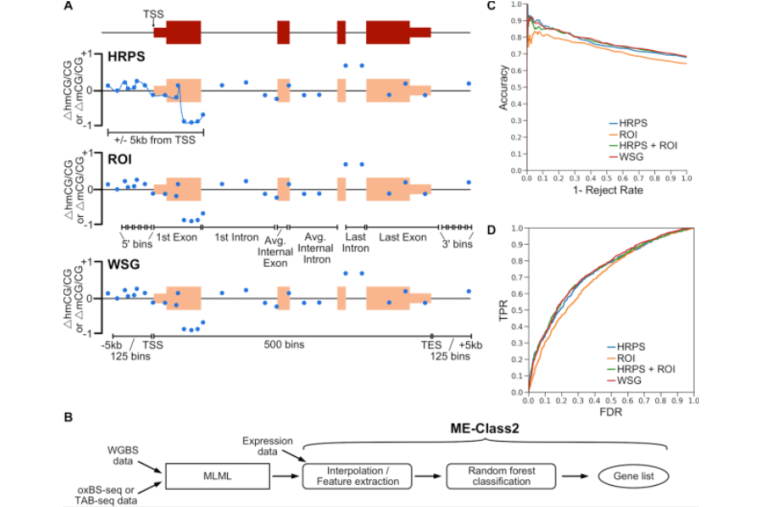
ME-Class2 reveals context dependent regulatory roles for 5-hydroxymethylcytosine
Schlosberg CE, Wu DY, Gabel HW, Edwards JR. Nucleic Acids Research. 18 March 2019. https://doi.org/10.1093/nar/gkz001
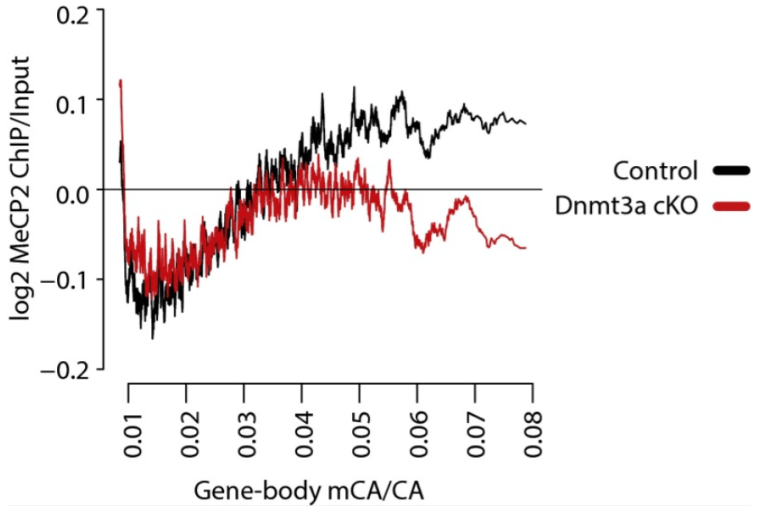
DNA methylation in the gene body influences MeCP2-mediated gene repression
Kinde BZ, Wu DY, Greenberg ME, Gabel HW. PNAS. 2016 Dec 27;113(52):15114-15119. doi: 10.1073/pnas.1618737114. Epub 2016 Dec 13.
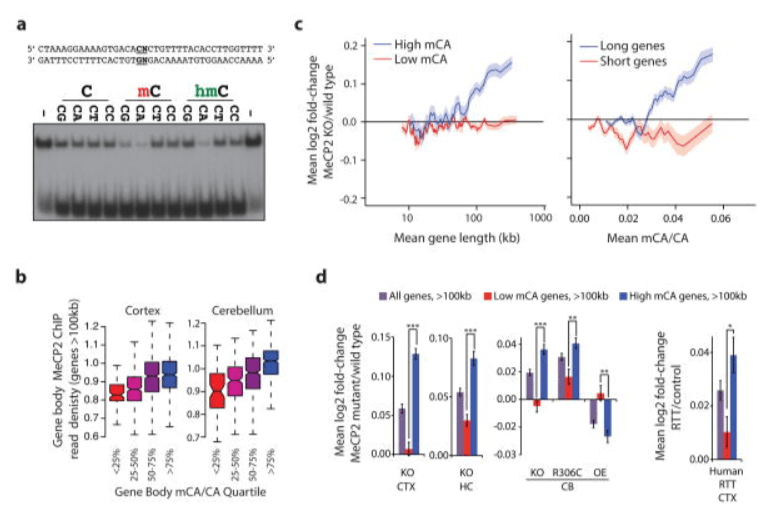
Disruption of DNA-methylation-dependent long gene repression in Rett syndrome
Gabel HW*, Kinde BZ*, Stroud H, Harmin DA, Hemberg M, Gilbert CS, Ebert DH, Greenberg ME. Nature. 2015 Jun 4;522(7554):89-93. doi: 10.1038/nature14319. Epub 2015 Mar 11.
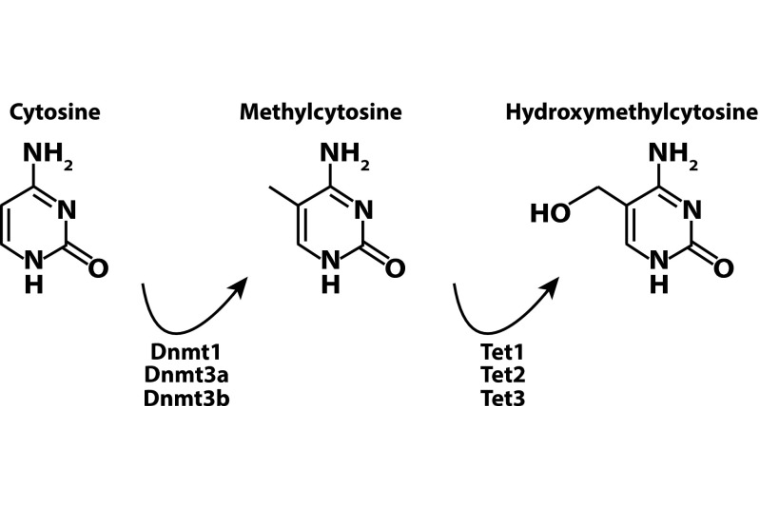
Reading the unique DNA methylation landscape of the brain: Non-CpG methylation, hydroxymethylation, and MeCP2
Kinde BZ*, Gabel HW*, Gilbert CS, Griffith EC, Greenberg ME. PNAS. 2015 Jun 2;112(22):6800-6. doi: 10.1073/pnas.1411269112. Epub 2015 Mar 4.
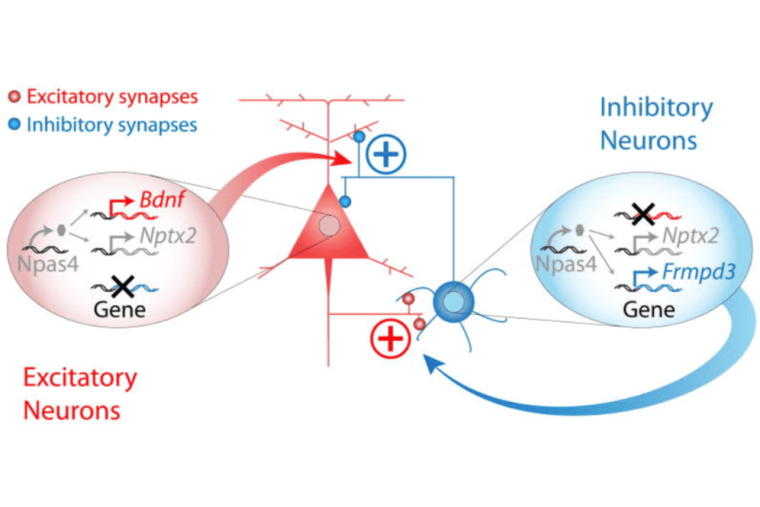
Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs
Spiegel* I, Mardinly* AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, Greenberg ME. Cell. 2014 May 22;157(5):1216-29. doi: 10.1016/j.cell.2014.03.058.

Activity-dependent phosphorylation of MeCP2 T308 regulates interaction with NCoR
Ebert DH, Gabel HW, Robinson ND, Kastan NR, Hu LS, Navarro AJ, Lyst MJ, Ekiert R, Bird AP, Greenberg ME. Nature. 2013 Jul 18;499(7458):341-5. doi: 10.1038/nature12348
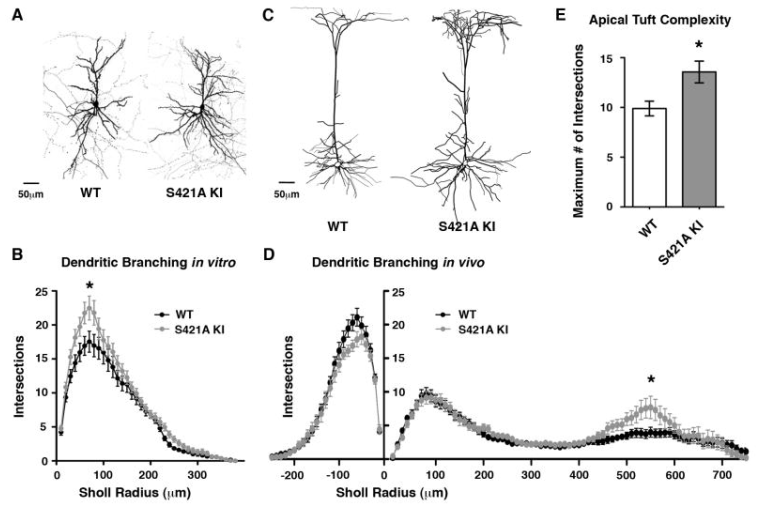
Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function
Cohen S*, Gabel HW*, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, Wetsel WC, West AE, Greenberg ME. Neuron. 2011 Oct 6;72(1):72-85.