The Dutcher Lab
The lab investigates the assembly and function of basal bodies/centrioles and cilia using genetics, biochemistry, microscopy, and computational biology in Chlamydomonas as well as human tissue culture cells. The key questions that we address are:
1.) how these microtubule-based structures are assembled,
2.) how they function and influence cellular biology, development, and human health.
Our interest in basal bodies/cilia led us to use bioinformatics and comparative genomics. We developed novel algorithms for identifying cilia and basal body/centriole genes. This study provided one of the first parts list for cilia and basal bodies. We identified the BBS5 gene, which is involved in ciliary signaling in a variety of cells and pathways in collaboration with many labs. This was the first example that the BBS gene affect cilia. Stemming from our comparative genomics work (Li et al. 2004), the discovery of many cilia and centriole-based diseases has illustrated the incredible breath of roles that these organelles play in human health. This work is now further studied using RNAseq (Albee et al., 2012) and proteomics (Lin et al., 2018; Bustamante et al., 2020). It appears that about 1,000 genes are needed to build functional motile cilia. We are now looking at the role of basal bodies in meiosis and the role of delta, epsilon and zeta tubulin.

One of the most important approaches for our studies is the use of microscopy. We have used electron tomography to study inner dyneins and basal body duplication while at the University of Colorado. Since coming to Washington University, we have used scanning disc confocal microscopy, quick freeze deep etch electron microscopy and single particle cryo electron microscopy to examine the transition zone (Lin et al., 2018), microtubule inner proteins (MIPs) (Ma et al., 2019) and the ciiary necklace (In preparation). With light microscopy, we have examined the asymmetric localization of a number of ciliary proteins (Ochi et al., 2020; Hwang et al., submitted). We are examining the role of several MIP proteins in determining the waveform (Bottier, Bayly, and Dutcher, in preparation).

Collaborations with labs in the Department of Mechanical Engineering has lead to biophysical understanding of the effects of ciliary length on waveform (Bottier et al., 2019) as well as develop acoustic traps to study wave form and cell-cell interactions (Kim et al., 2019; Cui et al., 2023).
Chlamydomonas reinhardtii

A very large family in Missouri, lead us to the assembly of cilia. This extended family had nine kids with primary ciliary dyskinesia that causes chronic respiratory infections, male infertility, and laterality defects. With exome capture, we identified the HEATR2 gene that is part of the cytoplasmic assembly pathway for building the axonemal dynein arms. We continue these studies on other assembly proteins using microscopy and genetics (Horani et al., 2023). We are currently studying proteins of a key heterodimer of CCDC39 and CCDC40 that is responsible for the placement of 100 proteins in motile airway cilia (Brody et al., in preparation).

For 2.5 years, I served as the Acting Director of the McDonnell Genome Institute (MGI),and become involved in many aspects of high-throughput genomics. While Director, I garnered a 10 million dollar contract for sequencing for the TOPMed project and wrote a 1.99 million dollar supplement to a grant to build better human and nonhuman primate references (Kronenber et al., 2018; Locke et al., 2019; Tallum et al., 2021).
We have used whole genome resequencing to find new mutants and suppressors of motility defects (Lin et al., 2015; Lin et al., 2018). Recently, we have used long range sequencing to obtain a telomere to telomere sequence of Chlamydomonas.
REPRESENTATIVE PUBLICATIONS (2018- PRESENT)
Chlamydomonas Biology
Lin, H., Zhang, Z. Iomini, C., and Dutcher, S.K. (2018). Identifying RNA splicing factors in Chlamydomonas reinhardtii. Open Biology, 8. pii: 170211. doi: 10.1098/rsob.170211.PMID:29514868
Hunter, E., Lechtreck, K., Hwang, J., Alford, L.M., Yamamoto, R., Kamiya, Lin, H., Yang, F., Dutcher, S.K., Wirschell, M. and Sale, W.S. (2018). IDA3 is a novel inner dynein arm assembly factor that is transported by IFT in growing cilia. MBoC, Feb 21. pii: mbc.E17-12-0729. doi: 10.1091/mbc.E17-12-0729. PMID:29467251.
Lin, H., Clifton, P., and Dutcher, S. K. (2018). High accuracy detection of insertional mutations in Chlamydomonas reinhardtii by MAPINS. Plant Physiol. 178: 1436-1447.
Lin, H., Guo, S., and Dutcher, S. K. (2018). RPGRIP1L helps to establish the ciliary gate for entry of proteins. J. Cell Sci., 131(20). pii: jcs220905. doi: 10.1242/jcs.220905.
Bottier M, Thomas KA, Dutcher SK, Bayly PV. (2019). How Does Cilium Length Affect Beating? Biophys J. 116(7):1292-304. Epub 2019/03/18. doi: 10.1016/j.bpj.2019.02.012. PMID: 30878201; Central PMCID: PMCPMC6451027.
Kim M, Huff E, Bottier M, Dutcher SK, Bayly PV, Meacham JM. (2019) Acoustic trap-and-release for rapid assessment of cell motility. Soft Matter. 15(21):4266-75. Epub 2019/04/11. doi: 10.1039/c9sm00184k. PMID: 30968924.
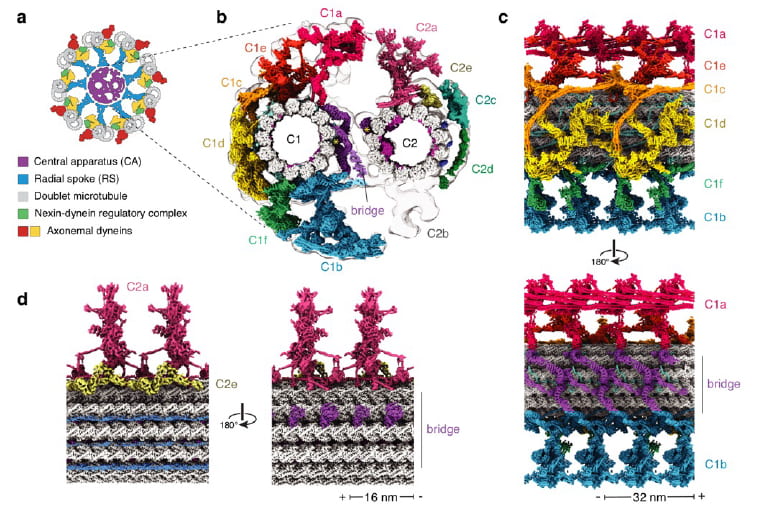
Ma M, Stoyanova M, Rademacher G, Dutcher SK, Brown A, Zhang R. (2019). Structure of the Decorated Ciliary Doublet Microtubule. Cell. 179(4):909-22 e12. Epub 2019/11/02. doi: 10.1016/j.cell.2019.09.030. PMID: 31668805; PMCID: PMC6936269.
Ochi, T., Quarantoitt, V., Lin, H., Jullien, J., Rosa e Silva, I., Boselli, F., Barnabas, D.D. Johnson, C. McLauglin, S. H., Freund, S. M. V., Blackford, A.N., Kimata, Y., Goldstein, R. E., Jackson, S. P., Blundell, T. L., Dutcher, S. K., Gergely, F., and van Breugel, M. (2020). CCDC61/VFL3 is a paralog of SAS6 and promotes ciliary functions. Structure, S0969-2126(20)30131-3. PMID: 32375023; PMCID: PMC7267773.
Pandey, M., Stormo, G.D., and Dutcher, S.K. (2020). Alternative Splicing during the Chlamydomonas reinhardtii Cell Cycle. G3, Oct 5;10(10):3797-3810. doi: 10.1534/g3.120.401622. PMID: 32817123; PMCID: PMC75334427.
Gui, M. Ma, M, Sze-Tu, E., Wang, X., Koh, Zhong, E., Berger, B., Davis, J., Dutcher, S., Zhang, R. and Brown, A. (2021). Structures of radial spokes and associated complexes important for ciliary motility, Nature Structural and Molecular Biology, doi: 10.1038/s41594-020-00530-0. PMID: 33318703 PMCID: PMC7855293.
Gui, M., Wang, X., Dutcher, S.K., Brown, A. and Zhang, R. (2022). Ciliary central apparatus structure reveals mechanisms of microtubule patterning. Nature Structural and Molecular Biology 29:483-492. PMID 35578023. Payne, Z. Penny, G., Turner, T. and Dutcher, S.K. (2022). A gap-free genome assembly of Chlamydomonas reinhardtii and detection of translocations induced by CRISPR-mediated mutagenesis. Plant Communicaions. v 17:100493. doi: 10.1016/j.xplc.2022.100493. PMID: 36397679.
Cui, M., Dutcher, S.K., Bayly, P.V. and Meacham, J. M. (2023). Robust acoustic trapping and perturbations of single cell micro-swimmers illuminate three-dimensional swimming and ciliary coordination. Proc. Natl. Acad. Sci, 120(25):e2218951120. doi: 10.1073/pnas.2218951120.
Collaborations on patient variants
Wambach, J.A. Wegner, D. J., Yang, P. Shinawi, M. Baldridge, D., Betleja, E. Shimonv, J., Hackett, B.P., Andrews, M.V., Ferkol, T., Dutcher, S.K., Mahjoub, M., and Cole, F. S. (2018). Functional Characterization of Biallelic RTTN Variants Identified in an Infant with Microcephaly, Simplified Gyral Pattern, Pontocerebellar Hypoplasia, and Seizures. PMID:29967526 DOI:10.1038/s41390-018-0083-z
Bustamante-Marin X. M., Horani A., Stoyanova, M. Charng, W.-L., Bottier, M., Sears, P. R., Yin, W.-L., Daniels, L. A., Bowen, H., Conrad, D.F. Knowles, M. R., Ostrowski, L. E., Zariwala, M. A., and Dutcher, S. K. (2020). Mutation of CFAP57, a protein required for the asymmetric targeting of a subset of inner dynein arms in Chlamydomonas, causes primary ciliary dyskinesia. PLoS Genetics, e1008691. doi: 10.1371/journal.pgen.1008691. eCollection 2020 Aug. PMID: 32764743.
Wallmeier J, Frank D, Shoemark A, Nothe-Menchen T, Cindric S, Olbrich H,. Loges, N.T., Gerard, I.A., Dougherty, W., Pennekamp, P., Kaiser, T., Mitchison, H.M., Hogg, C., Carr, S.B., Zariwala, M. A., Ferkol, T., Leigh, M.W., Davis, S.D., Atkinson, J., Dutcher, S.K., Knowles, M.R., Thiele, H., Altmuller, J., Krenz, H., Woste, M., Brentrup, A., Ahrens, F., Vogelberg, C., Morris-Rosendahl, D.J. and Omran, H. (2019). De Novo Mutations in FOXJ1 Result in a Motile Ciliopathy with Hydrocephalus and Randomization of Left/Right Body Asymmetry. Am J Hum Genet. 2019;105(5):1030-9. Epub 2019/10/22. doi: 10.1016/j.ajhg.2019.09.022. PMID: 31630787; PMCID: PMC6849114.
Horani, A., Gupta, D.K., Xu, J., Xu, H., Puga-Molina, L.C., Santi, C.M. Ramagiri, S. Brennen, S.K., Pan, J., Huang, T. Hyland, R.M., Gunsten, S.P., Tzeng, S.C., Strahle, J.M., Mill, P., Mahjoub, M.R., Dutcher, S.K., Brody, S.L. (2023).. The effect of Dnaaf5 gene dosage on primary ciliary dyskinesia phenotypes, JCI insights, PMID: 36712068
A Few From Time at MGI and Genomics (2018- present)
Kronenberg, Z. N., Fiddes, I.T., Gordon, G., Murali, S., Cantsilieris, S., Meyerson, O.S., Underwood, J.G., Nelson, B.J., Chaisson, M.J.P., Dougherty, M.L., Munson, K.M., Hastie, A., Diekhans, M., Hormozdiari, F., Lorusso, N., Hoekzema, K., Qiu, R., Clark, K., Raja, A., Welch, A.E., Sorensen, M., Baker, C., Fulton, R.S., Armstrong, J., Graves-Lindsay, T.A., Denli, A.M., Hoppe, E.R., Hill, C.M., Pang, A.W.C., Lee, J., Lam, E.T., Dutcher, S.K., Gage, F.H., Warren, W.C., Shendure, J., Haussler, D., Schneider, V.A., Cao, H., Ventura, M., Wilson, R.K., Paten, B., Pollen, A., and Eichler, E.E. (2018). High
resolution comparative analysis of great ape genomes. Science. PMID:29880660 DOI:10.1126/science.aar6343.
Locke AE, Steinberg KM, Chiang CWK, Service SK, Havulinna AS, Stell L, et al. (2019) Exome sequencing of Finnish isolates enhances rare-variant association power. Nature. 572(7769):323-8. Epub 2019/08/02. doi: 10.1038/s41586-019-1457-z. PMID: 31367044; Central PMCID: PMC6697530.
Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, Taliun SAG, Corvelo A, Gogarten SM, Kang HM, Pitsillides AN, LeFaive J, Lee SB, Tian X, Browning BL, Das S, Emde AK, Clarke WE, Loesch DP, Shetty AC, Blackwell TW, Smith AV, Wong Q, Liu X, Conomos MP, Bobo DM, Aguet F, Albert C, Alonso A, Ardlie KG, Arking DE, Aslibekyan S, Auer PL, Barnard J, Barr RG, Barwick L, Becker LC, Beer RL, Benjamin EJ, Bielak LF, Blangero J, Boehnke M, Bowden DW, Brody JA, Burchard EG, Cade BE, Casella JF, Chalazan B, Chasman DI, Chen YI, Cho MH, Choi SH, Chung MK, Clish CB, Correa A, Curran JE, Custer B, Darbar D, Daya M, de Andrade M, DeMeo DL, Dutcher SK, Ellinor PT, Emery LS, Eng C, Fatkin D, Fingerlin T, Forer L, Fornage M, Franceschini N, Fuchsberger C, Fullerton SM, Germer S, Gladwin MT, Gottlieb DJ, Guo X, Hall ME, He J, Heard-Costa NL, Heckbert SR, Irvin MR, Johnsen JM, Johnson AD, Kaplan R, Kardia SLR, Kelly T, Kelly S, Kenny EE, Kiel DP, Klemmer R, Konkle BA, Kooperberg C, Köttgen A, Lange LA, Lasky-Su J, Levy D, Lin X, Lin KH, Liu C, Loos RJF, Garman L, Gerszten R, Lubitz SA, Lunetta KL, Mak ACY, Manichaikul A, Manning AK, Mathias RA, McManus DD, McGarvey ST, Meigs JB, Meyers DA, Mikulla JL, Minear MA, Mitchell BD, Mohanty S, Montasser ME, Montgomery C, Morrison AC, Murabito JM, Natale A, Natarajan P, Nelson SC, North KE, O’Connell JR, Palmer ND, Pankratz N, Peloso GM, Peyser PA, Pleiness J, Post WS, Psaty BM, Rao DC, Redline S, Reiner AP, Roden D, Rotter JI, Ruczinski I, Sarnowski C, Schoenherr S, Schwartz DA, Seo JS, Seshadri S, Sheehan VA, Sheu WH, Shoemaker MB, Smith NL, Smith JA, Sotoodehnia N, Stilp AM, Tang W, Taylor KD, Telen M, Thornton TA, Tracy RP, Van Den Berg DJ, Vasan RS, Viaud-Martinez KA, Vrieze S, Weeks DE, Weir BS, Weiss ST, Weng LC, Willer CJ, Zhang Y, Zhao X, Arnett DK, Ashley-Koch AE, Barnes KC, Boerwinkle E, Gabriel S, Gibbs R, Rice KM, Rich SS, Silverman EK, Qasba P, Gan W; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium, Papanicolaou GJ, Nickerson DA, Browning SR, Zody MC, Zöllner S, Wilson JG, Cupples LA, Laurie CC, Jaquish CE, Hernandez RD, O’Connor TD, Abecasis GR. (2021). Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. Feb;590(7845):290-299. doi: 10.1038/s41586-021-03205-y. Epub 2021 Feb 10. PMID: 33568819; PMCID: PMC7875770.