Molecular genetic analysis of plant-pathogen interactions:
Understanding pathogen virulence and host susceptibility
My research group is interested in the regulatory mechanisms governing interactions between bacterial plant pathogens and their hosts. Specifically, we would like to understand the molecular basis of pathogenesis and disease development, and are focusing on the identification and characterization of both pathogen virulence factors and their targets in plant cells. My group studies the bacterial plant pathogen Pseudomonas syringae and two of its hosts, Arabidopsis thaliana (a small weed in the mustard family) and tomato, systems in which both pathogen and host are amenable to genetic and molecular analyses. We are using a combination of molecular and genetic approaches to identify both pathogen and plant genes that govern development of disease. These studies have led to important insights regarding the mechanisms of virulence and indicate that pathogens manipulate normal physiological processes in plants to promote disease.
How does the pathogen gain the upper hand?
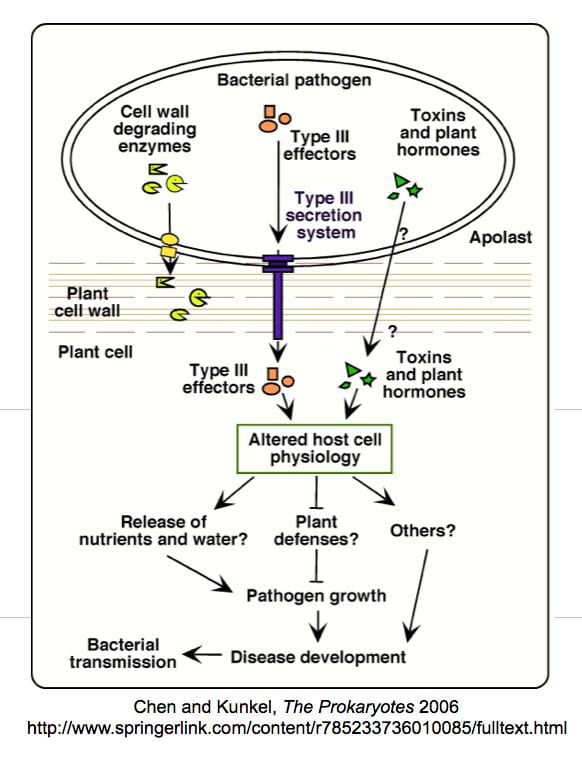
P. syringae grows to high levels in leaves, in the spaces between plant cells, a region referred to as the “apoplast”. To render plant tissue suitable for microbial growth pathogens must alter the physiology of the host. Such modifications include inhibition of anti-microbial defenses, release of water and/or nutrients into the apoplast, and development of disease symptoms. Previous studies by others have revealed that P. syringae utilizes at least two different mechanisms to deliver virulence factors that promote these events: i) secretion of toxins into the apoplast and ii) direct injection of bacterial proteins into the host cell through a specialized delivery apparatus known as the Type III secretion system (TTSS). Although several P. syringae toxins have been characterized, their modes of action and their targets in the plant are not well understood. Likewise, several proteins known to be injected into plant cells via the P. syringae TTSS have recently been identified, but their role in promoting disease is unclear.
The overall goal of my research program is to identify and characterize P. syringae virulence factors and to gain a better understanding of their mode of action within plant cells. By using a combination of bacterial genetics to identify P. syringae virulence factors and plant genetics to dissect the basis of disease susceptibility in A. thaliana (and thus learn something about the mode of action of virulence factors) we are in a unique position to study the mechanisms underlying pathogenesis and disease development in this system. Our studies should also provide insight into the molecular basis of disease development in other plant-pathogen interactions, a contribution that could result in the development of improved strategies for preventing or controlling plant disease in agricultural and horticultural systems.
Identification and characterization of P. syringae virulence factors How does the pathogen gain the upper hand?
We have been using two complementary approaches to identify and characterize P. syringae genes involved in pathogenesis. The first involves a genetic screen for P. syringae mutants that are impaired in their ability to cause disease on A. thaliana and tomato. The second approach is to investigate the mechanism by which a P. syringae protein, AvrRpt2, that known to be injected into plant cells contributes to virulence. The mutant screen resulted in the identification of several new virulence factors, including the transcriptional regulator TvrR (Preiter et al, 2005) and the tricarboxylic acid cycle enzyme Mqo (Mellgren et al, 2009), as well as the isolation of several new mutants that disrrupt the synthesis of the phytotoxin, coronatine (COR; Brooks et al, 2004), which is a molecular mimic of the plant hormone JA-Ile.

The phytotoxin coronatine plays three distinct roles during pathogenesis. The screen for P. syringae mutants with reduced virulence resulted in identification of both previously known (e.g. toxin biosynthesis, type III secretion components, etc) and several potential new virulence genes. One of the most exciting results of this screen was the isolation of a series of mutants defective for synthesis of the P. syringae toxin coronatine (COR; Brooks et al, 2004). COR has been shown to be important for P. syringae virulence in several plant species, including A. thaliana and tomato. The chemical structure of COR resembles the plant signaling molecule JA-Ile, an amino acid conjugate of jasmonic acid.
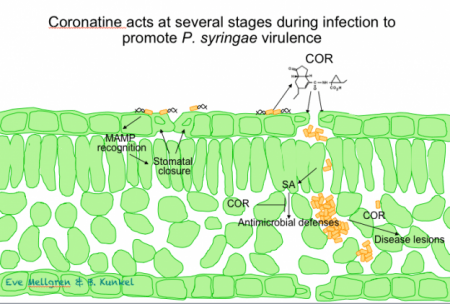
We and others have shown that COR promotes disease by modulating jasmonate signaling within the plant. Specifically, COR promotes disease through at least three different mechanisms: 1) promoting entry of P. syringae into the leaf tissue by re-opening of stomates (Melotto et al, 2006), 2) promoting growth in the apoplast by suppressing Salicylic acid (SA)-mediated defenses (Brooks et al, 2005), and 3) promoting diease lesion development via an SA-independent mechanism. These studies are nicely complemented by our studies on the role of jasmonate signaling within the host (see below). Our COR biosynthetic mutants ( Brooks et al, 2004) are freely available to the community and provide a valuable tool for future studies investigating the role of COR in pathogenesis of A. thaliana and tomato.
The P. syringae AvrRpt2 Type III Secreted effector protein alters host auxin physiology. The P. syringae AvrRpt2 protein is injected directly into plant cells via the Type Three Secretion System (TTSS). It has been proposed that the primary function of effector proteins such as AvrRpt2 is to promote pathogen virulence, and this has been demonstrated for several TTSS-secreted proteins. However, the function of these proteins, and the mechanisms by which they contribute to disease development are not well understood. We have shown that AvrRpt2 functions as a virulence factor on both A. thaliana and tomato plants (Chen et al, 2000; Lim and Kunkel, 2005). Using transgenic A. thaliana plants that produce the AvrRpt2 protein, we have demonstrated that this bacterial protein functions inside the plant to promote pathogen growth and development of disease (Chen et al, 2000). This was the first time a type III-injected protein was shown to promote virulence from within a plant cell. Our more recent findings suggest that AvrRpt2 functions either downstream or independently of salicylic acid to promote pathogen virulence (Chen et al, 2004). Thus, AvrRpt2 may promote pathogen virulence via a mechanism that does not involve suppression of known host defenses.
Insert AvrRpt2 vir activity in planta, Chen 2007.
AvrRpt2 is a cysteine protease with several demonstrated proteolytic targets within plant cells (Chisholm et al, 2005; PNAS 102:2087), including the A. thaliana protein RIN4. One or more of these host proteins are proposed to be virulence targets of AvrRpt2. We and others have demonstrated, however, that the virulence activity of AvrRpt2 is not dependent upon cleavage and/or degradation of RIN4 (Lim & Kunkel, 2004b). We are currently utilizing physiological, genetic and molecular approaches to investigate the mechanism(s) by which AvrRpt2 promotes P. syringae pathogenesis.
Analysis of disease susceptibility in A. thaliana
To complement our work on P. syringae virulence factors we are using plant genetics to identify targets of virulence factors in the host. This has primarily involved looking for A. thaliana mutants that are less susceptible to P. syringae infection. This approach has provided valuable insight regarding the mechanisms by which pathogen virulence factors alter physiological processes in the host to promote disease. An example in which our analysis of disease susceptibility in A. thaliana has converged with our studies of pathogen virulence is the isolation and characterization of COR-insensitive A. thaliana mutants. Our analysis of these mutants has provided insight regarding the mechanisms by which COR alters host normal host cell biology to promote pathogen virulence, and suggests that COR may function in two ways to promote pathogenesis: i) by inhibiting anti-microbial defense responses in the plant and ii) by promoting disease symptom development (Kloek et al, 2001; Laurie-Berry et al, 2006).
Our studies on the mode of action of the P. syringea virulence factor AvrRpt2 in the plant indicates that auxin physiology of the host may be an important virulence target. To learn more about this we are also investigating the role of auxin during pathogenesis.
Investigating the role of the plant hormone auxin in P. syringae Pathogenesis.
Details coming soon.